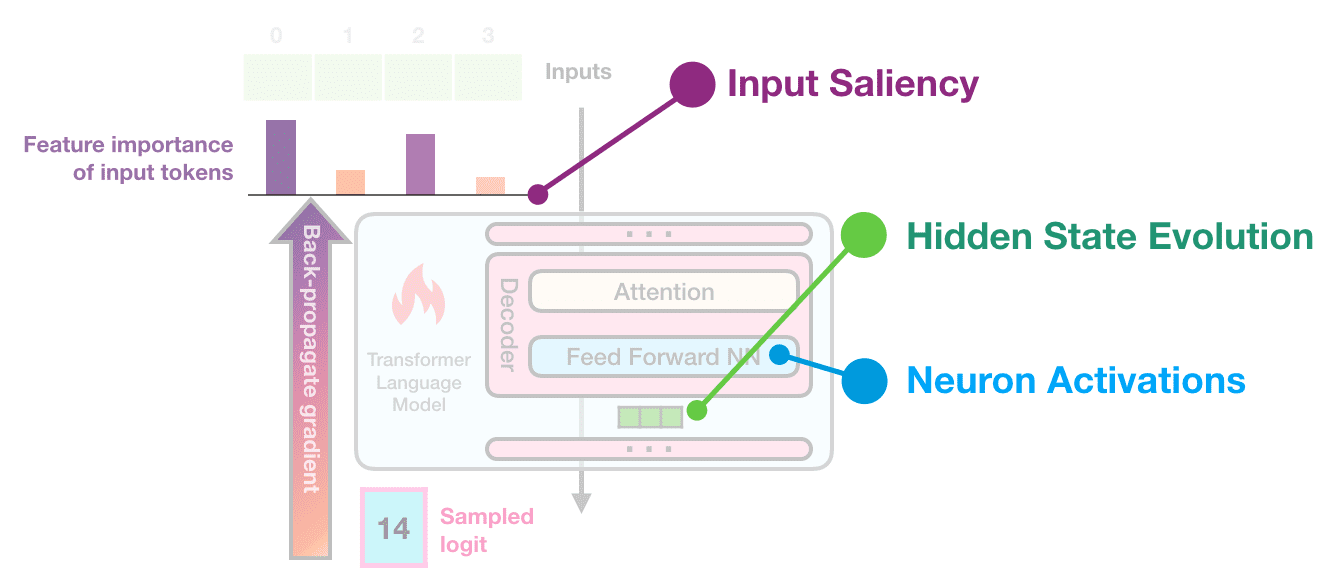
Final DXA-nation
Longitudinal Image-based AI Models for Health and Medicine
AI can see the end! Deep learning predicts all-cause mortality from single and sequential body composition imaging

Key Points, TLDR:
- The combination of body composition imaging and meta-data (e.g. age, sex, grip strength, walking speed, etc) resulted in the best 10 year mortality predictions
- Longitudinal or sequential models overall performed better than single record models, highlighting the importance of modeling change and time dependencies in health data.
- Longitudinal models have the potential to provide a more comprehensive assessment of one’s health
- Read the paper
Artificial intelligence (AI) and machine learning (ML) are revolutionizing healthcare, driving us toward the era of precision medicine. The motivation to develop AI health models is to reduce deaths and disease as well as prolong a high quality of life. Well trained models have the ability to more thoroughly analyze data that is presented which offers a more comprehensive assessment of one’s health.
Single Record vs Longitudinal Models
Image-based medical AI/ML models have now reached a maturity where they often rival or even surpass human performance, adeptly identifying patterns and anomalies that could easily elude the human eye. However, the majority of these models still operate on single time-point data, providing an isolated snapshot of health at one specific instance. Whether these are uni-modal or multi-modal models, they tend to work with data gathered within a relatively similar timeframe, forming the foundation of a prediction. Yet, in the broader context of AI/ML for medical applications, these single time-point models represent just the first step — the proverbial ‘low hanging fruit.’ One frontier of medical AI research is longitudinal models which offer a more holistic view of a person’s health over time.
Longitudinal models are designed to integrate data from multiple time-points, capturing an individual’s health trajectory rather than a standalone moment. These models tap into the dynamic nature of human health, where physiological changes are constant. The ability to map these changes to specific outcomes or health questions could be a game-changer in predictive healthcare. The concept of longitudinal data isn’t new to clinical practice — it’s regularly used to monitor aging and predict frailty. A prime example is the tracking of bone mineral density (BMD), a key marker for osteoporosis and frailty. Regular assessments of BMD can detect significant decreases, indicating potential health risks.
Longitudinal Model Development Challenges
Historically, the development of longitudinal models has faced several significant challenges. Aside from larger data volumes and computation required per individual, the most critical obstacle lies in the curation of longitudinal medical data itself. Unlike single time-point data, longitudinal data involves tracking patients’ health information over prolonged periods, often across multiple healthcare institutions. This requires meticulous data organization and management, making the curation process both time-consuming and expensive. Multiple successful studies have been funded to prospectively collect longitudinal data. These studies report challenges with respect to patient retention over a longer observation period. Hence, despite the potential benefits of longitudinal models, their development has remained a complex, resource-intensive endeavor.
The Goal
Changes in body composition, proportions of lean and fat soft tissue and bone, are known to be associated with mortality. In our study, we aimed to use body composition information to better predict all-cause mortality, in simpler terms, the likely timeline of a person’s life. We evaluated the performance of models built on both single time-point and longitudinal data, respectively referred to as our ‘single record’ and ‘sequential’ models. Single record models allowed us to evaluate what type of information was most predictive of mortality. Development of sequential models were for the purposes of capturing change over time and evaluating how that affects mortality predictions.
The Data
The data for this study was acquired from a longitudinal known as the Health, Aging, and Body Composition (Health ABC) study in which over 3000 older, multi-race male and female adults were followed and monitored for up to 16 years. This study resulted in a rich and comprehensive longitudinal data set. As a part of this study patients received total body dual energy X-ray absorptiometry (TBDXA) imaging and several pieces of meta-data were collected (see table XXX). Consistent with best modeling practices and to avoid data leakage or mitigate overfitting, the data was split into a train, validation, and hold-out test set using a 70%/10%/20% split.
We quantify body composition using total body dual energy X-ray absorptiometry (TBDXA) imaging which has long been considered a gold standard imaging modality. Historically, patient meta-data which include variables like age, body mass index (BMI), grip strength, walking speed, etc were used to assess aging/mortality and used as surrogate measurement of body composition. The prevalent use of patient meta-data and surrogate measures of body composition were driven by the limited accessibility to DXA scanners. Accessibility has improved greatly as of recent with scans becoming cheaper and no longer needing a physician referral/order/prescription.
Single Image Model
Three single record models were built each with different data inputs but all with the same output which was a 10 year mortality probability. The first model was built to only take patient meta-data and is a neural network with a single 32-unit, ReLU activation hidden layer and sigmoid prediction layer. The second model used only TBDXA images as input and it consisted of a modified Densenet121 which was modified to handle the two color channels as opposed to three color channels (RGB) seen in most natural images. The dual energy nature of DXA results in a high and low X-ray images which are fully registered and stacked into two image channels. The third model combines the meta-data embedding of model one with the TBDXA image embeddings of model two then passes it through a 512-unit, a 64-unit fully-connected ReLU layer to make, and lastly a sigmoid prediction layer.

Longitudinal/Sequential Model
Three sequential models were built and evaluated. The single record model architectures served as the base for each sequential model but the sigmoid prediction layers were removed so that the output was a vector representing feature embeddings. Over the course of the study data was collected from each patient at multiple time points. The data from each time point was input into the appropriate models to acquire the corresponding feature vector. The feature vectors for each patient were ordered and stacked into a sequence. A Long Short Term Memory (LSTM) model was trained to take the sequence of feature vectors and output a 10 year mortality prediction. As previously mentioned, there are several difficulties with conducting long term studies with retention and data collection being a common problem. Our study was not absent of these problems and some patients had more data points that others as a result. An LSTM model was chosen as the sequence modeling approach because they are not constrained to use the same input sequence length for each patient. I.e. LSTMs can work with sequences of varying length thus eliminating the need to pad sequences if patients were short the full set of data points (~10).

Image + Meta-data Longitudinal Models Win
Area under the receiver operating characteristic (AUROC) on the hold-out test set show that metadata performs better than using TBDXA image alone in both the single record and sequential models. However, combining meta-data and TBDXA imaging resulted in the best AUROCs in both modeling paradigms which indicates that imaging contains useful information, predictive of mortality that is not captured by the meta-data. Another way to interpret this is that the meta-data are not a full surrogate measure of body composition with respect to predicting mortality. If they were full surrogates, combining TBDXA imaging with meta-data would have resulted in no significant increase or change in AUROC. The fact that the combination resulted in better AUROCs indicates that imaging is providing orthogonal information beyond what the meta-data capture and further justifies the utility of imaging.

Longitudinal or sequential models overall performed better than single record models. This is true across all modeling approaches and input data types (meta-data, image only, combined meta-data and image). These results demonstrate the importance of modeling change and the time dependencies of health data.
We performed an Integrated Discrimination Improvement (IDI) analysis to evaluate the benefits of combining imaging with metadata, compared to using metadata alone. This analysis was conducted on the sequence models, which outperformed the single-record models. The IDI was found to be 5.79, with an integrated sensitivity and specificity of 3.46 and 2.33, respectively. This indicates that the combination of imaging and metadata improves the model’s ability to correctly identify those who will not survive the next 10 years by 3.46%, and enhances the ability to correctly identify those who will survive the next 10 years by 2.33%. Overall, this suggests an improvement in model performance of approximately 5.8%.

So What?
Our study underscores the promising potential of longitudinal AI/ML models in the realm of predictive healthcare, specifically in the context of all-cause mortality. The comparative analysis of single record models and longitudinal models revealed that the latter offers superior performance, indicating the critical role of modeling change over time in health data analysis. The clinical implication of our findings include the ability to provide a more precise and holistic assessment of one’s health through models that account for a patient’s historical or longitudinal data. While the data needed for developing longitudinal health models exists, the proper infrastructure and institutional support is not quite oriented yet to enable efficient data curation and development of these models at scale. Nevertheless, many are working to overcome these hurdles and the development of longitudinal models is one of many exciting frontiers for AI in medicine.
The clinical implications of these findings are far-reaching. Longitudinal models have the potential to transform care delivery by enabling more precise, personalized predictions about a patient’s health trajectory. Such models can inform proactive interventions, thereby enhancing care outcomes and possibly even prolonging life. Moreover, the use of both metadata and imaging data sets a new precedent for future AI/ML models, suggesting a synergistic approach for optimal results. It reinforces the need for multidimensional, nuanced data to paint an accurate and holistic picture of a patient’s health. These findings represent significant strides in the application of AI/ML in healthcare, highlighting an exciting path forward in our pursuit of precision medicine.
More Resources:
Read the paper
More Information about IDI and published examples:
- Area Under the Curve and Beyond
- Dual-energy three-compartment breast imaging for compositional biomarkers to improve detection of malignant lesions – Communications Medicine
- GitHub – LambertLeong/AUC_NRI_IDI_python_functions: Custom python functions to help you further analyse machine learning models and diagnostic test
- Area Under the Curve and Beyond with Integrated Discrimination Improvement and Net Reclassification
Final DXA-nation was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.